Research led by Yashar Zeighami at McGill University, Canada, provides a new way to characterize brain diseases. The study, published in the open access journal PLOS Biology on April 20 shows that comparing the transcriptomes (the map of activity for all genes in the genome) related to different brain diseases can help us understand the mechanisms underlying the diseases, and why certain ones are comorbid. The method can also find new relationships among diseases, which could have an impact on clinical treatment options.
Classifying brain diseases is difficult because many have multiple genetic and environmental risk factors. In addition, the symptoms of many brain diseases overlap. For example, Parkinson’s disease and dementia with Lewy bodies are both neurodegenerative disorders presenting with muscle tremors and rigidity, and which share some similar cognitive and behavioral symptoms.
In this and other similar cases, misdiagnosis is not uncommon and can have serious consequences for patient care. An alternative approach is to classify brain diseases based on gene activity. The new study examined disease transcriptomes—the set of RNA transcripts from affected brain regions—for 40 different brain diseases.
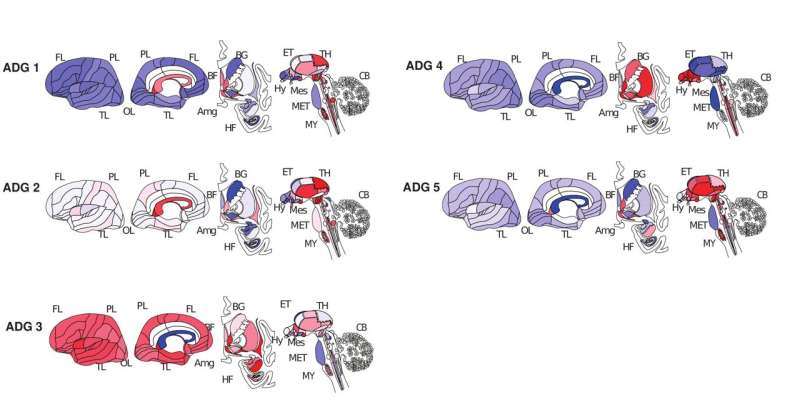
The researchers found that this system could classify brain diseases into five primary groups based on where disease-risk genes were active in the brain and in which cell types. In addition to confirming known relationships among diseases, disease transcriptome analysis was able to find previously unknown relationships among diseases.
For example, language development disorders, obsessive-compulsive disorder, and temporal lobe epilepsy were all classified into group 3, meaning that despite their very different symptoms, their corresponding genes are active in the same brain regions and in the same cell types.
Brain diseases classified as neurodegenerative, movement-related, and psychiatric are the most difficult to diagnose because of the overlap in symptoms that change over time. The transcriptome is thus an additional tool that could be used for more accurate early diagnoses.
Zeighami adds, “Analysis of the transcription patterns of risk genes for human brain disease reveals characteristic expression signatures across brain anatomy. These can be used to compare and aggregate diseases, providing associations that often differ from conventional phenotypic classification.”
More information:
A comparison of anatomic and cellular transcriptome structures across 40 human brain diseases, PLOS Biology (2023). DOI: 10.1371/journal.pbio.3002058
Journal information:
PLoS Biology
Source: Read Full Article