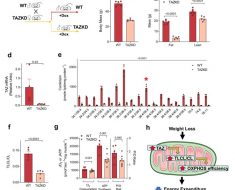
Researchers have genetically engineered the first mice that get a human-like form of COVID-19, according to a study published online November 1 in Nature.
Led by researchers from NYU Grossman School of Medicine, the new work created lab mice with human genetic material for ACE2—a protein snagged by the pandemic virus so it can attach to human cells as part of the infection. The mice with this genetic change developed symptoms similar to young humans infected with the virus causing COVID-19, instead of dying upon infection as had occurred with prior mouse models.
“That these mice survive creates the first animal model that mimics the form of COVID-19 seen in most people—down to the immune system cells activated and comparable symptoms,” said senior study author Jef Boeke, the Sol and Judith Bergstein Director of the Institute for Systems Genetics at NYU Langone Health. “This has been a major missing piece in efforts to develop new drugs against this virus.”
“Given that mice have been the lead genetic model for decades,” added Boeke, “there are thousands of existing mouse lines that can now be crossbred with our humanized ACE2 mice to study how the body reacts differently to the virus in patients with diabetes or obesity, or as people age.”
Problem of large DNA
The new study revolves around a new method to edit DNA, the 3 billion “letters” of the genetic code that serve as instructions for building our cells and bodies.
While famous techniques like CRISPR enable the editing of DNA editing just one or a few letters at a time, some challenges require changes throughout genes that can be up to 2 million letters long. In such cases, it may be more efficient to build DNA from scratch, with far-flung changes made in large swaths of code pre-assembled and then swapped into a cell in place of its natural counterpart.
Because human genes are so complex, Boeke’s lab first developed its “genome writing” approach in yeast, one-celled fungi that share many features with human cells but that are simpler and easier to study.
More recently, Boeke’s team has adapted their yeast techniques to the mammalian genetic code, which is made up of not just of genes that encode proteins, but also of many switches that turn on different genes at different levels in different cell types.
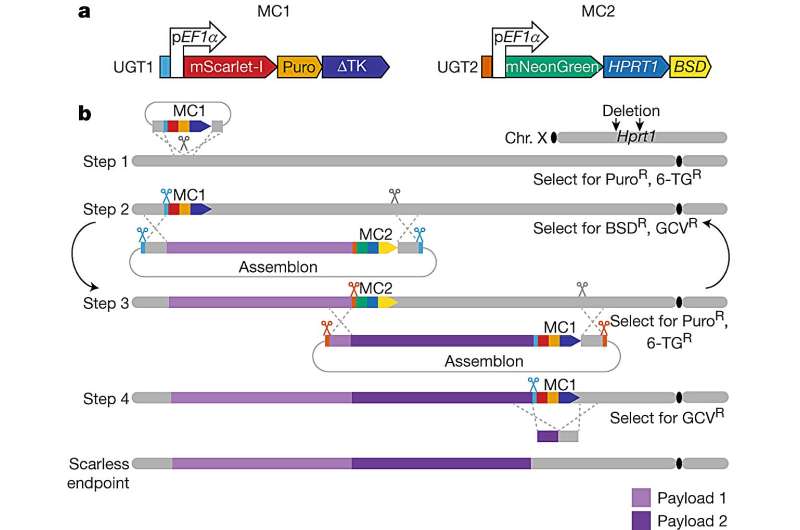
By studying this poorly understood “dark matter” that regulates genes, the research team was able to design living mice with cells that had more human-like levels of ACE gene activity for the first time. The study authors used yeast cells to assemble DNA sequences of up to 200,000 letters in a single step, and then delivered these “naked” DNAs into mouse embryonic stem cells using their new delivery method, mSwAP-In.
Overcoming the size limits of past methods, mSwAP-In delivered a humanized mouse model of COVID-19 pathology by “overwriting” 72 kilobases (kb) of mouse Ace2 code with 180 kb of the human ACE2 gene and its regulatory DNA.
To accomplish this cross-species swap, the study method cut into a key spot in the DNA code around the natural gene, swapped in a synthetic counterpart in steps, and with each addition, added a quality control mechanism so that only cells with the synthetic gene survived. The research team then worked with Sang Yong Kim at NYU’s Rodent Genome Engineering Lab using a stem cell technique called “tetraploid complementation” to create a living mouse whose cells included the overwritten genes.
In addition, the researchers had previously designed a synthetic version of the gene Trp53, the mouse version of the human gene TP53, and swapped it into mouse cells. The protein encoded by this gene coordinates the cell’s response to damaged DNA, and can even instruct cells containing it to die to prevent the build-up of cancerous cells. When this “guardian of the genome” itself becomes faulty, it is a major contributor to human cancers.
Whereas the ACE2 experiments had swapped in an unchanged version of a human gene, the synthetic, swapped-in Trp53 gene had been designed to no longer include a combination of molecular code letters—cytosine (C) next to guanine (G)—known to be vulnerable to random, cancer-causing changes. The researchers overwrote key CG “hotspots” with code containing a different DNA letter in adenine (A).
“The AG switch left the gene’s function intact, but lessened its vulnerability to mutation, with the swap predicted to lead to a 10-to-50 fold lower mutation rate,” said first author Weimin Zhang, Ph.D., a post-doctoral scholar in Boeke’s lab. “Our goal is to demonstrate in a living test animal that this swap leads to fewer mutations and fewer resulting tumors, and those experiments are being planned.”
Along with Boeke and Zhang, NYU Langone study authors were Ran Brosh, Aleksandra Wudzinska, Yinan Zhu, Noor Chalhoub, Emily Huang, and Hannah Ashe in the Institute for Systems Genetics and Department of Biochemistry & Molecular Pharmacology; Ilona Golynker, Lucia Carrau, Payal Damani-Yokota, Camille Khairallah, Kamal Khanna, and Benjamin tenOever in the Department of Microbiology; and Matthew Maurano and Sang Yong Kim in the Department of Pathology.
More information:
Jef Boeke, Mouse genome rewriting and tailoring of three important disease loci, Nature (2023). DOI: 10.1038/s41586-023-06675-4. www.nature.com/articles/s41586-023-06675-4
Journal information:
Nature
Source: Read Full Article