Earlier this month, biologist Mohammed AlQuraishi was driven to excitedly exclaim that a new set of findings constituted “a seismic and unprecedented shift so profound it has literally turned a field upside down over night.” You don’t read that every day.
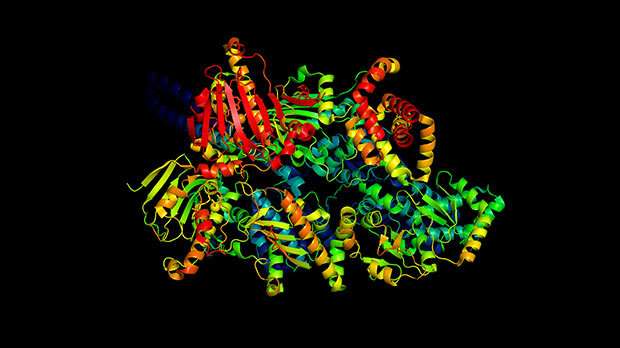
What had led him to make such a bold claim? It was the news that Google-owned British artificial intelligence company, DeepMind, had ‘cracked’ the decades-old conundrum of how proteins fold with a new version of their deep learning system, AlphaFold.
Proteins are fundamental to all life and the mysterious way they fold into elaborate 3-D shapes has dramatic implications for how our cells and tissues operate, so this was certainly big news and the headlines echoed as much. But what did the findings tell us, what could they mean for cancer research and was the hyperbole justified? We asked two of our expert protein specialists, who are trying to understand more about how the way proteins fold affect cancer outcomes, to give their verdict on the news.
What problem has DeepMind solved?
The ‘protein folding problem’ has remained a headscratcher since it was first posed around 50 years ago. In a nutshell, being able to predict how a protein folds is key to understanding how it will function, which in turn could unlock answers to big questions, including how to treat diseases like cancer.
Researchers have put a great deal of time, effort and resource into trying to scrutinize the way proteins fold, which have led to the emergence of innovative but expensive experimental techniques to study protein structures such as X-ray crystallography, which creates a 3-D structure of a protein using, you guessed it, X-ray beams. There are other techniques too, such as using an electron microscope to beam electrons onto a protein to magnify its image.
But while these are considered optimal techniques, they each have their limitations. X-ray crystallography only really works when studying stable proteins that can form the neat crystals required for the process. And even then, it’s a laborious and costly task. With flexible or ‘unstable’ proteins, which have less structure and rigidity, it’s a whole other ball game.
But luckily, back in 1963, American biochemist Christian Anfinsen proposed that a protein’s one dimensional sequence of amino acids—something far easier to obtain—should give away its full 3-D structure. This work earned him a Nobel Prize in 1972. And since that time, scientists have been exploring this route using less expensive and more accessible computational methods. But therein lies another problem. There are almost infinite numbers of ways that a protein could fold and identifying them all can take a lifetime. Not so good for tackling important challenges like cancer.
This is where DeepMind comes in. Their AlphaFold technology uses deep learning to take what we already know about the structures of known proteins held on a global database and ‘learn’ about and therefore predict the structures of other proteins. The team tested the predictions made by AlphaFold against structures that had been determined experimentally, using methods like X-ray crystallography. And the results looked extremely promising. AlphaFold managed to predict the structures of proteins with a never-before-seen level of accuracy at low cost and within days, not years or decades. And this is what got the scientific community just a tiny bit excited.
So what do our protein experts think?
Professor Richard Bayliss at the University of Leeds uses crystallography to determine the shape of proteins and how they fold. This knowledge is vital to uncovering their function in cancer cells and importantly, how they can be targeted and treated. His particular focus is on the Myc protein, which is associated with many different cancers, including aggressive forms of prostate and breast cancers.
Dr. Patricia Muller at our Cancer Research UK Manchester Institute is researching a protein that plays an important role in stopping cancer developing, the p53 protein. She’s particularly interested in how p53 functions in both its unfolded and folded states and believes that the latter has an impact on how it operates in cancers.
What did you make of the news? Is it as monumental as the headlines would have us believe?
Richard: It’s certainly an impressive and exciting advance, but in a more limited and specific sense than some of the headlines would lead you to believe. Being able to accurately predict the structure of a protein from its amino acid sequence has been something of a holy grail for structural biologists. The work from DeepMind is the first time that this has been achieved with a level of reliability that can compete with the experimental methods. It’s enough to give you the confidence to make substantial investment in the predictions that arise from the structure.
It’s limited in several ways, but perhaps the most important is that it cannot accurately predict the structure of proteins that work intimately with other proteins, and whose structure is therefore dependent on the presence of these other proteins. This sort of problem is a major focus of structural biologists, and the structures predicted by DeepMind will be useful in helping us to interpret and use experimental data.
Patricia: Our understanding of protein folding mainly comes from studies using techniques that take a long time and a lot of patience. If AlphaFold is as accurate as the articles suggest, it would really make a big difference and speed up many different lines of research.
What could this mean for cancer research?
Richard: Despite the recent technological advances in experimental techniques, such as cryogenic electron microscopy and high-throughput crystallography, determining a protein structure is a challenge that can take many years to solve. Some projects are held up at the very first step because the protein of interest cannot be made in sufficient quantity or purity for the methods to work, or they turn out to be unstable proteins. This is where having a reliable computational method could provide enough information to advance a project. For example, if we want to map the location of cancer mutations onto the structure of the protein to predict how they might affect its function, we could do this much more quickly with the DeepMind computational method than experimental methods.
It’s difficult to say how useful it will be for drug discovery, which is an important application of structural biology in cancer research, because we depend on accurate models generated by X-ray crystallography. But sometimes we have to use computational models for proteins with unknown structures and having more reliable models will help us to develop drugs more efficiently.
Patricia: It could help in many ways, such as developing drugs or simply understanding more about how proteins work. As an example, some drugs work by preventing proteins from binding to other proteins. If we know their structure, we can predict how they will bind to each other and we can then design drugs that would prevent the binding between these proteins. For the proteins that we do know the structure of, this has been shown to be a successful strategy. However, the techniques that we have for elucidating protein structure don’t work for every protein. Predictions from AlphaFold could be very useful for determining the structures of proteins that other techniques have not yet worked for.
How will you use this new information to advance your work?
Richard: I study the Myc protein, which is associated with many human cancers but is regarded as “undruggable” by most researchers because it doesn’t have a fixed structure. The DeepMind approach will not be much help for Myc itself because it only adopts a structure when it binds to other proteins.
But it could be very useful to study the binding proteins that partner with Myc, some of which are very challenging proteins to work with. We don’t expect Myc to affect the structures of its binding partners, and so predictions made by AlphaFold about these other proteins are likely to be accurate and useful.
These structures, together with our experimental data, would enable us to generate models of the interactions between Myc and these partner proteins. And this information could be used to develop molecules that block the interactions, which we see as a route to developing new cancer treatments.
Patricia: My work is on the p53 protein. We don’t yet know the full structure of p53 and it’s very difficult to determine its shape with conventional techniques. Currently, we rely on antibodies to help us detect the structure, but they can only detect two states: folded and unfolded. But I believe there’s more to the story than that and that there are actually several different states of folded p53. I would love to see if AlphaFold could help me uncover these states because in cancer, there are hundreds of different ways that p53 mutates. I believe the technology could help me reveal why this is the case and shed light on how these mutants are different from each other.
So what have we learned?
Source: Read Full Article