Nanobodies have different mechanisms to neutralize viruses, and some target epitopes not accessible to monoclonal antibodies, making them an alternative for developing therapies for coronavirus disease 2019 (COVID-19).
.jpg)
Neutralizing monoclonal antibodies are one form of therapy being developed for use against COVID-19, with some having received emergency approval. Apart from this, antibody fragments or nanobodies have also been developed for neutralizing viruses. They are smaller than monoclonal antibodies, but can still bind viruses with great specificity.
Since they are small, they can be designed in various forms to bind different virus epitopes, preventing virus mutational escape. They are generally stable and soluble and can be produced in microbes such as yeasts and E. coli, quickly and easily. They can also be made into aerosols for delivery via inhalation.
In the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, the receptor binding (RBD) domain of the virus spike protein binds to the angiotensin-converting enzyme 2 (ACE2) of the host. Because most neutralizing antibodies are directed to the RBD, it is also a region where several mutations have arisen, reducing the potency of monoclonal antibodies.
Previously, researchers reported several nanobodies that can bind to the RBD. In a new paper published in the bioRxiv* preprint server, researchers report the effect of the different RBD mutations on the potency of these nanobodies.
Nanobodies have different mechanisms of neutralization
The researchers found that six different RBD mutations had a negligible effect on most of the nanobodies, except for the E484K mutation, which completely reduced the affinity of two nanobodies.
Using pseudoviruses, they also evaluated the effect of the new variants B.1.1.7 initially found in the United Kingdom and the South African B.1.351 strains. The B.1.1.7 strain having the RBD mutation N501Y did not have much effect on the nanobodies. The B.1.351 variant with three RBD mutations reduces the effect of two nanobodies, but does not affect other nanobodies much. In contrast, most monoclonal antibodies and convalescent and vaccine-elicited antibodies are greatly affected by at least one of these strains.
The authors used cryo-electron microscopy (EM) to understand the structural reasons for the potency of these nanobodies. The images of the nanobody complexes with the spike protein or RBD revealed three separate classes of nanobodies that were affected differently.
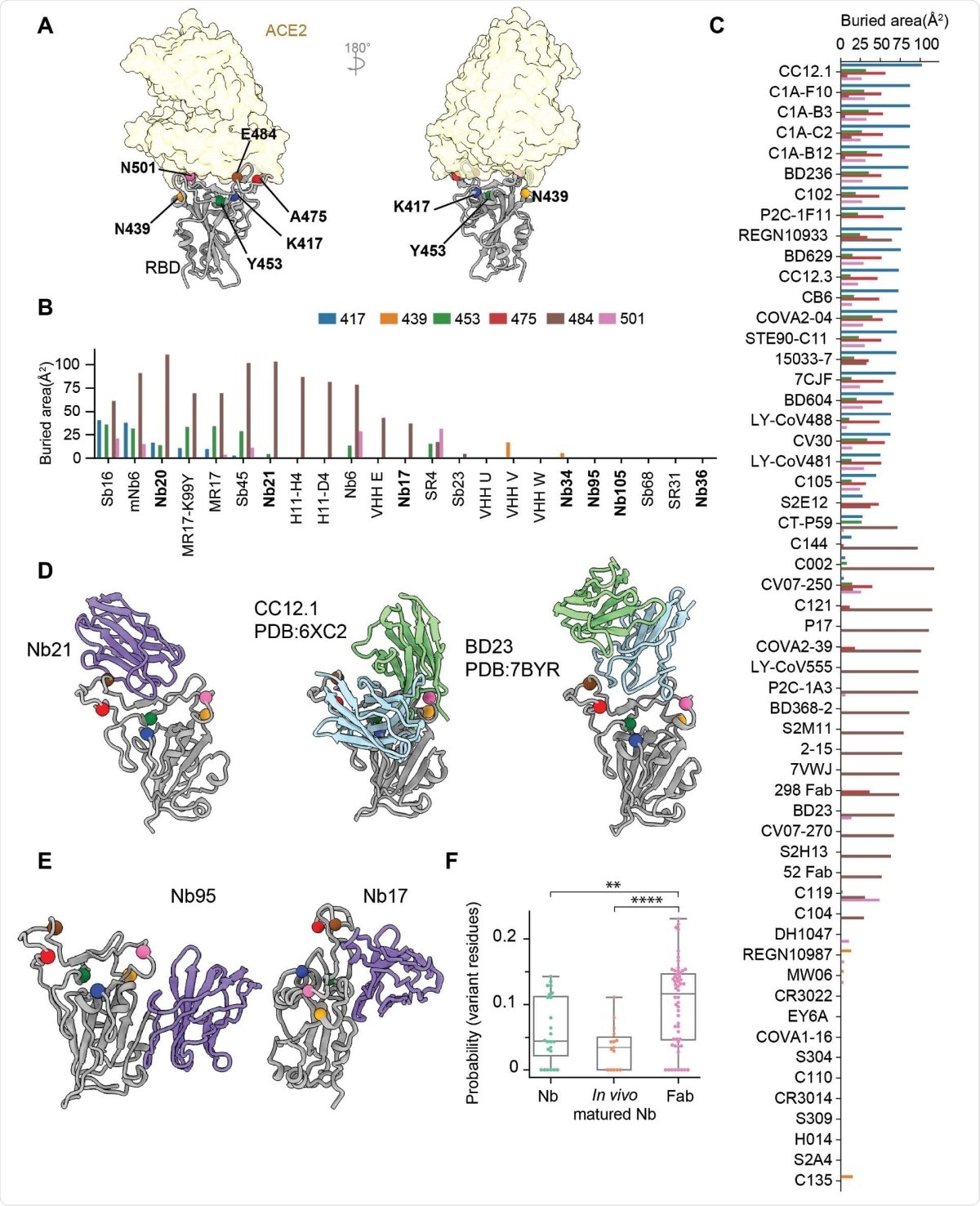
Class I nanobodies, having short CDR3 residues, represent some of the most potent SARS-CoV-2 neutralizers. The cryo-EM structures revealed that the most potent Class I nanobody binds the RBD in both the up and down conformation. However, the E484K can significantly reduce the binding of this nanobody by destabilizing the interface packing that helps bind the nanobody to the RBD.
Class II nanobodies can neutralize the virus at a higher concentration (below 150 ng/ml) than the Class I nanobodies (below 15 ng/ml). They bind to the RBD mainly via hydrophobic interactions. Although they do not interact with ACE2 directly, they can still block ACE2 binding at low enough concentrations.
One of the Class III nanobodies, Nb17, is extremely effective at neutralizing SARS-CoV-2 in vitro. Because of its high affinity to the spike protein, it can lock the S1 subunit into a rigid and open conformation that prevents fusion to the host cell membrane. Nb17 is also resistant to all the RBD mutations seen so far. Another Class III nanobody can destabilize the spike protein and binds the RBD in a manner that is different from the binding of Nb17.
More potent than antibodies
The binding of Class III nanobodies is new and different from the binding of Class I and Class II nanobodies, which have common epitopes with monoclonal antibodies. Class III nanbodies have epitopes close to the N-terminal domain, which are difficult to access for the monoclonal antibodies because of their large size. Because Nb17 binds to the RBD-up conformations, it could potentially be used along with vaccines to expose conserved RBD epitopes that could elicit a broad antibody response.
The majority of the serum antibodies target RBD residues; the RBD mutations are effective means to escape neutralization. Since most monoclonal antibodies are derived from convalescent plasma, they become less potent against new variants. This is different from nanobodies that have never co-evolved with the virus and hence are less sensitive to mutations.
Nanobodies being derived from camelids or identified using in vitro affinity studies, along with their smaller structures, can target conserved and unique epitopes on the RBD that cannot be accessed by monoclonal antibodies. Thus, such nanobodies can be used to design better COVID-19 therapies and vaccines.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Sun, D. et al. (2021) Potent neutralizing nanobodies resist convergent circulating variants of SARS-CoV-2 by targeting novel and conserved epitopes. bioRxiv. https://doi.org/10.1101/2021.03.09.434592, https://www.biorxiv.org/content/10.1101/2021.03.09.434592v1
Posted in: Medical Science News | Medical Research News | Disease/Infection News | Healthcare News
Tags: ACE2, Angiotensin, Angiotensin-Converting Enzyme 2, Antibodies, Antibody, Cell, Cell Membrane, Convalescent Plasma, Coronavirus, Coronavirus Disease COVID-19, E. coli, Electron, Electron Microscopy, Enzyme, in vitro, Microscopy, Mutation, Nanobodies, Protein, Receptor, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Vaccine, Virus

Written by
Lakshmi Supriya
Lakshmi Supriya got her BSc in Industrial Chemistry from IIT Kharagpur (India) and a Ph.D. in Polymer Science and Engineering from Virginia Tech (USA).
Source: Read Full Article





