A new study presents an innovative approach to the crucial detection of pre-cancerous lesions using large, high-res images. A team of researchers from Portugal developed a machine learning solution that assists pathologists in the detection of cervical dysplasia, making the diagnosis of new samples completely automatic. It’s one of the first published works to use full slides.
Cervical cancer is the fourth most frequent cancer among women, with an estimated 604,000 new cases in 2020, according to the World Health Organization (WHO). However, it is also among the most successfully preventable and treatable types of cancer, provided it is early identified and properly managed. Hence, screening and detection of pre‑cancerous lesions (and vaccination) are crucial to prevent the disease.
But what if we could develop machine learning models to help the subjective classification of lesions in the squamous epithelium—the type of epithelium that has protective functions against microorganisms—using whole‑slide images (WSI) containing information from the entire tissue.
In this sense, a team of researchers from the Institute for Systems and Computer Engineering, Technology and Science (INESC TEC) and from the molecular and anatomic pathology laboratory IMP Diagnostics, in Portugal, developed a weakly‑supervised methodology—a machine learning technique that combines annotated and non-annotated data during model training—to grade cervical dysplasia.
This is particularly useful, given that pathology data annotations are difficult to obtain: the images are huge, which makes the annotation process very time-consuming and tedious, in addition to its high subjectivity. This type of technique allows researchers to develop models with good performance, even with some missing information during the model training phase.
The model will then grade cervical dysplasia, the abnormal growth of cells on the surface, as low (LSIL) or high-grade intraepithelial squamous lesions (HSIL).
“In the detection of cervical dysplasia, this was one of the first published works that use the full slides, following an approach that includes the segmentation and subsequent classification of the areas of interest, making the diagnosis of new samples completely automatic,” explained Sara Oliveira, a researcher at INESC TEC.
This process of classification is complex and can be subjective. Therefore, the development of machine learning models can assist pathologists in this task; moreover, computer-aided diagnosis (CAD) plays an important role: these systems can serve as a first indication of suspicious cases, alerting pathologists to cases that should be more closely evaluated.
Sara Oliveira reinforced that even the development of CAD systems for decision support in digital pathology is far from being completely solved. “In fact, computational pathology is still a relatively recent area, with many challenges to solve, so that machine learning models can effectively approach clinical applicability,” she mentioned.
There’s also a compromise at play in using WSI, and the most common approaches focus on the manual clipping of smaller areas of the slides. WSI are usually large, high-resolution images (often larger than 50,000 × 50,000 pixels); therefore, they’re not easily adaptable to the graphics processing units (GPU) used to train deep learning models.
“Despite promising results, the fact that these approaches require manual selection of the areas to be classified, focusing only on small regions (taking into account the size of the slide), makes them more fragile from an implementation point of view,” said the researcher.
Training the segmentation model
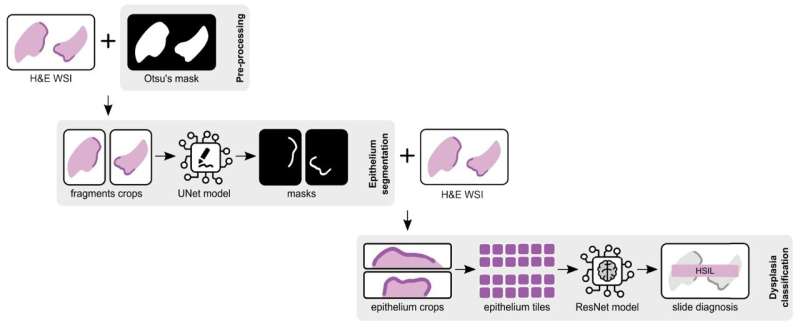
The framework comprises an epithelium segmentation step followed by a dysplasia classifier (non‑neoplastic, LSIL, HSIL), making the slide assessment completely automatic, without the need for manual identification of epithelial areas. “The proposed classification approach achieved a balanced accuracy of 71.07% and sensitivity of 72.18%, at the slide‑level testing on 600 independent samples,” clarified the lead author of the study.
To train the segmentation model, the researchers used all the annotated slides (186), with a total of 312 tissue fragments. The results show that “only very rarely does the model fail to recognize a large part of the epithelium or misidentify a significant area.”
After the first step of segmentation, the researchers used the identified ROIs to focus on for the classification, allowing the use of non-annotated WSI for training, and the automatic diagnosis of unseen cases. Then, the classifier can diagnose the dysplasia grade from tiles of those areas.
This solution used 383 annotated epithelial regions to train the classification model, divided into training and validation sets. The researchers tested different models and, after choosing the best one, in an attempt to leverage the classification learning task, they re-trained the version by adding some individual labeled tiles to the training set (263). By combining the selected tile of each epithelium area, that only has the label of the correspondent bag, with tiles that have a particular label associated, the tile selection process was improved.
Finally, to take advantage of the complete dataset, the team re-trained the model by adding bags of tiles from the non-annotated slides (1198).
The lead researcher of the paper reinforces that future work could aim to refine both parts of the model (segmentation and classification), as well as evaluate a fully integrated approach.
The test set of 600 samples, used in the current study, was selected from the IMP Diagnostics dataset and is available “upon reasonable request.”
“At IMP Diagnostics we are invested in improving cervical cancer diagnosis and, thus, women’s health. This tool is a step closer to a more efficient detection of pre-malignant lesions,” concludes Diana Montezuma Felizardo, Pathologist and Head of R&D at the IMP Diagnostics.
The findings are published in the journal Scientific Reports.
More information:
Sara P. Oliveira et al, A CAD system for automatic dysplasia grading on H&E cervical whole-slide images, Scientific Reports (2023). DOI: 10.1038/s41598-023-30497-z
Journal information:
Scientific Reports
Source: Read Full Article